 |
IMC is characterized by being able to perform operations of cloning experiment on computer. At this time, special data is unnecessary, it is possible to clone base sequence data that can be obtained from a public database such as GenBank or EMBL as it is. For cloning, restriction enzyme digestion, PCR primer design, PCR amplification, and ligation can be performed without changing the annotated sequence. All resulting cloning products are output in GenBank / EMBL format. Since Primer information is pasted and stored on the base sequence, it is also useful for Primer management.
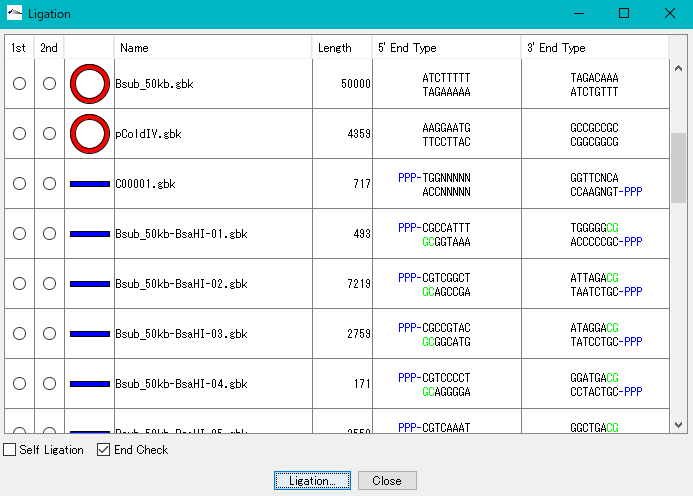
- • It is a cloning function that can actually be disconnected / ligated.
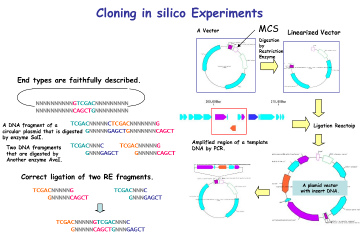
- • In silico cloning experiments are possible.
- • It helps to understand invisible molecular biology experiments. It can be used for assisting and simulating molecular biology experiments.
- • You can construct arbitrary Vector / Plasmid.
- • List restriction enzymes that are most suitable for insert checks on Vector and simulate the gel electrophoresis results.
- • Restriction enzyme treatment (restriction enzyme map, restriction enzyme digestion fragment generation, optimal restriction enzyme candidate list for insert check, others)
- • PCR primer design,
- • Primer design to avoid certain features,
- • Replication by PCR (including notes),
- • Batch primer design to amplify all genes,
- • Primer management ligation, self-ligation,
- • Base fragment end shape conformity check
- • Plasmid map creation (insert area blowing function) (corresponding to layout style)
- • Addition of a restriction enzyme recognition site to the cloning DNA end with the annotation described
- • DNA terminal blunting, phosphorylation (dephosphorylation
- • Cutting arbitrary region of annotated base sequence
|